Enumeration And Size Distribution Of Yeast Cells In The Brewing Industry
Breweries, such as those operated by Anheuser Busch, Inc., utilize the Coulter Principle for their yeast cell enumeration and sizing. This technology uses the Z™ Series instrument models and the Multisizer 3. This document will focus on the Z Series models: the Z1 Single Threshold, the Z1 Dual Threshold, and the Z2 Analyzer.
Yeast cells are analyzed during the fermentation process. Carbohydrates in the wort, which is the water containing sugars from the grains, are converted by the yeast strain into alcohol, carbon dioxide, and numerous by-products.
There are several stages in this process, during which the yeast cells are analyzed. The first, and very critical stage for cell enumeration is during the pitching process. Pitching is the introduction of the live yeast culture into cooled wort to begin the fermentation. It is important to deliver the proper cell concentration for a successful fermentation. In addition, the yeast concentration will affect beer taste. A typical pitch concentration is 30.3 x 106 cells/mL. Yeast counts using either a Z1 single or dual threshold can be used to determine the initial cell concentration. If size distribution is being measured, a Z2 model is necessary.
Often, samples are taken during the fermentation process. During this time, the yeast cells may be budding. The Z Series instruments will enumerate a budding cell as a single unit. The third stage for sampling is at the end of the fermentation process. A sample analyzed at this time is highly likely to have proteinaceous material present. To accurately measure these samples, a Z2 analyzer is needed. The size-distribution histogram may show a bi-modal size population. One peak is the yeast cell population; the other, flocculated protein (Figure 1). To remove the protein material, simply add several drops of 2 M sodium hydroxide (NaOH) to the yeast sample, mix, and analyze on the Z2.
Equipment
One of the following instruments
- Z1 Single Threshold P/N 6605698
- Z1 Dual Threshold P/N 6605699
- Z2 Analyzer P/N 6605700
- Accuvette™ Cups BCI P/N 8320592
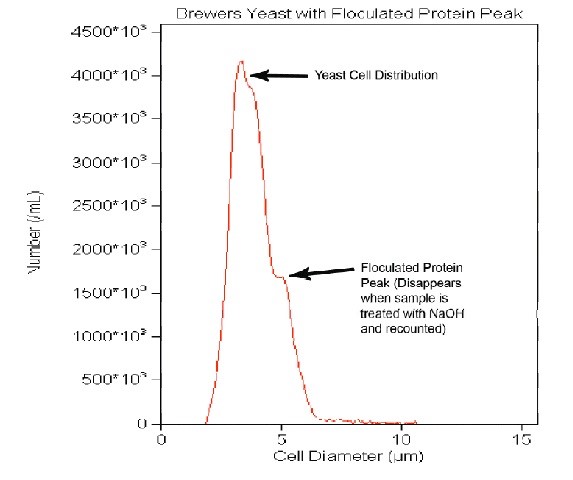
Figure 1: A yeast sample with a bi-modal peak. The first peak is yeast, while the second peak is that of flocculated protein. The secondary peak can be larger than the yeast cell peak.
Reagents
- 2 M NaOH Sigma P/N S8263
- Z pak BCI P/N 8320312
- L3 Beads BCI P/N 6602793
- L5 Beads BCI P/N 6602794
- L10 Beads BCI P/N 6602796
Method
Basic Instrument Setup for Z1 Dual Threshold and Z2 Analyzer
- Sample preparation: 10 µL of yeast sample into 20 mL Isoton II
- 100 micron Aperture Tube.
- Set Lower Threshold to 3.0 microns.
- Set Upper Threshold to 10.0 microns.
- Count Mode - between TL and TU.
- Set Dilution Factor 2000.
Basic Instrument Setup for Z1 Single Threshold
- Sample preparation: 10 µL of yeast sample into 20 mL Isoton II
- 100 micron Aperture Tube.
- Set Lower Threshold to 3.0 microns.
- Count Mode - above TL.
- Set Dilution Factor 2000.
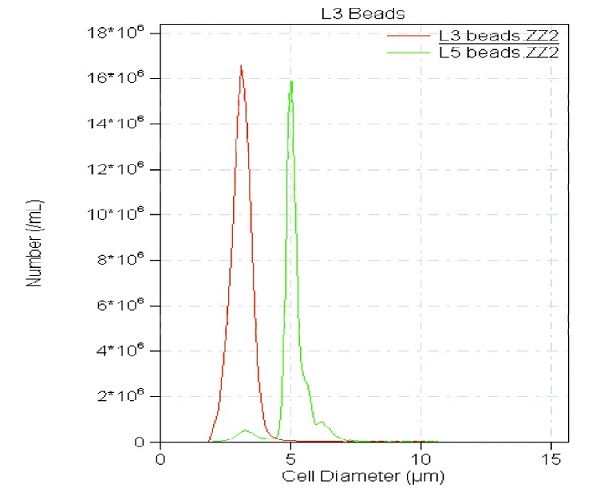
Figure 2: L3 and L5 beads used to set size parameters
Procedure
- Calibrate Z system. Add 5 drops of L10 Beads to 20 mL of Isoton II.
- Mix gently by inversion.
Note: Do not mix vigorously as air bubbles will form. - Press Cal button on the keypad to select the C1 page.
- Set units to µm. In the calibration size, enter Number Mode.
- Press Start.
- Record calibration factor.
- Repeat 3 times.
- Average results and enter in new calibration factor.
- Mix gently by inversion.
- Running size standard
- Select Set Up.
- Set lower threshold to 3.0 microns.
- Set Upper Threshold to 10 microns.
- Set count mode to between.
- Add 5 drops of L5 Beads to 20 mL of Isoton II.
- Mix by inversion.
- Press start.
- After run, acquire data with Accucomp software (Figure 2).
- If there are sample controls, run these at this
- Run samples
- Select output.
- Set dilution factor to 2000. Note: Dilution factor may vary based on sample density.
- Set result type to concentration.
- Add 10 µL of sample to 20 mL Isoton II. Note: Sample may have aggregate; mix thoroughly prior to adding to Isoton II.
- Mix gently.
- Press start.
- After run, acquire data with Accucomp™ software (Figure 3).
- Samples that have a bi-modal peak (Figure 1).
- Add 4 drops of 2 M NaOH.
- Mix gently.
- Incubate 5 min. and Press start.
- After run acquire data with Accucomp software.
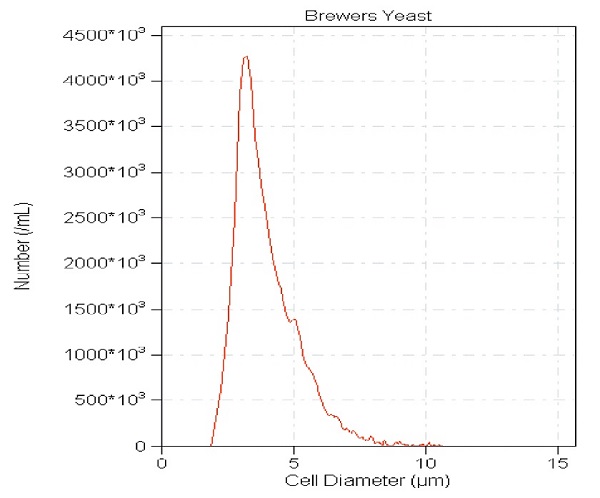
Figure 3: Brewers Yeast, which was grown at 37ºC 2 hours in distilled water. Average yeast cell size is 3.5 microns. Aggregates show as 7 microns or higher
Selected References
- ASTM D F2149-01. Standard Test Method for Automated Analyses of Cells-the Electrical Sensing Zone Method of Enumerating and Sizing Single Cell Suspensions. ASTM
International. February, 2002. - H.A. Teass, Jr., J. Byrnes & A. Valentine. Correlation of Yeast Measurement Between Spin Down and Coulter Counter Laboratory Analysis and In-Line Measurements With the McNab Cell Counter. Master Brewers Association of the Americas Volume 35, Number 2, 1998 Pages 101-103.
* All trademarks are the property of their respective owners.
Helpful Links
-
阅读材料
-
应用手册
- 17-Marker, 18-Color Human Blood Phenotyping Made Easy with Flow Cytometry
- 21 CFR 第 11 部分关于在线 WFI 仪器的数据完整性要求
- 8011+ Reporting Standards Feature and Synopsis
- Air Particle Monitoring ISO 21501-4 Impact
- Automated Cell Plating and Growth Assays
- Automated Cord Blood Cell Viability and Concentration Measurements Using the Vi‑CELL XR
- Automated Genomic Sample Prep RNAdvance
- Biomek Automated NGS Solutions Accelerate Genomic Research
- Biomek i-Series Automation of the Beckman Coulter GenFind V3 Blood and Serum DNA Isolation Kit
- Preparation and purification of carbon nanotubes using an ultracentrifuge and automatic dispensing apparatus, and analysis using an analytical centrifuge system
- Viability Assessment of Cell Cultures Using the CytoFLEX
- Classifying a Small Cleanroom using the MET ONE HHPC 6+
- Clean Cabinet Air Particle Evaluation
- Recommended cleaning procedure for the exterior surface of the MET ONE 3400+
- Counting Efficiency: MET ONE Air Particle Counters and Compliance to ISO-21501
- Critical Particle Size Distribution for Cement using Laser Diffraction
- CytoFLEX
- Detecting and counting bacteria with the CytoFLEX research flow cytometer: II-Characterization of a variety of gram-positive bacteria
- Efficient kit-free nucleic acid isolation uses a combination of precipitation and centrifugation separation methods
- Compensation Setup For High Content DURAClone Reagents
- Echo System-Enhanced SMART-Seq v2 for RNA Sequencing
- Grading of nanocellulose using a centrifuge
- Grading of pigment ink and measurement of particle diameter using ultracentrifugation / dynamic light scattering
- A Highly Consistent Bradford Assay on Biomek i-Series
- How to Use Violet Laser Side Scatter Detect Nanoparticle
- HRLD Recommended Volume Setting
- Particle Size Analysis Simple, Effective and Precise
- Flow Cytometric Analysis of auto-fluorescent cells found in the marine demosponge Clathria prolifera
- MET ONE Sensor Verification
- Metal colloid purification and concentration using ultracentrifugation
- Separation and purification of metal nanorods using density gradient centrifugation
- Miniaturized and High-Throughput Metabolic Stability Assay Enabled by the Echo Liquid Handler
- Minimal Sample to Sample Carry Over with the HIAC 8011+
- Modern Trends in Non‐Viable Particle Monitoring during Aseptic Processing
- Particle diameter measurement of a nanoparticle composite - Using density gradient ultracentrifugation and dynamic light scattering
- Identification of Circulating Myeloid Cell Populations in NLRP3 Null Mice
- Optimizing the HIAC 8011+ Particle Counter for Analyzing Viscous Fluids
- Particle testing in cleanroom high-pressure gas lines to ISO 14644 made easy with the MET ONE 3400 gas calibrations
- Pharma Manufacturing Environmental Monitoring
- Pharma Manufacturing Paperless Monitoring
- Analysis of plant genome sizes using flow cytometry: a case study demonstrating dynamic range and measurement linearity
- Protein purification workflow
- Calibrating the QbD1200 TOC Analyzer
- Detection Limit
- USP 787 Small Volume Testing
- A fully automated plate-based optimization of fed-batch culture conditions for monoclonal antibody-producing CHO cell line
- A Deeper Look at Lipid Nanoparticles
- A High-Throughput, Automated Screening Platform for IgG Quantification During Drug Discovery and Development
- Automated Research Flow Cytometry Workflow Using DURA Innovations Dry Reagent Technology with the *Biomek i7 Automated Workstation and *CytoFLEX LX Flow Cytometer
- Automating antibody titration using a CytoFLEX LX analyzer Integrated with a Biomek i7 Multichannel workstation and Cytobank streamlined data analysis
- Automated IDT Alt-R CRISPR/Cas9 Ribonucleoprotein Lipofection Using the Biomek i7 Hybrid Automated Workstation
- Monitoring Plant Cell Cultures with BioLector and Multisizer 4e Instruments
- Monitoring Yeast Cultures with the BioLector and Multisizer 4e instruments
- Biomek i7 Hybrid Automated KAPA mRNA HyperPrep Workflow
- Comparative Analysis of DURAClone IM T-Cell Subsets Antibody Panel: Conventional vs. Spectral Flow Cytometry on CytoFLEX mosaic Spectral Detection Module
- Cultivation of suspended plant cells in the BioLector®
- CytoFLEX mosaic Spectral Detection Module Enables Enhanced Spectral Unmixing of White Blood Cell Populations by Extracting Multiple Autofluorescence Signatures
- 利用BioLector进行细胞死亡的测定
- Echo System-Enhanced SMART-Seq v4 for RNA Sequencing
- A Simple Guide to Selecting the Right Handheld Particle Counter for Monitoring Controlled Environments
- Linearity of the Vi-CELL BLU Cell Counter and Analyzer
- Miniaturized 16S rRNA Amplicon Sequencing with the Echo 525 Liquid Handler for Metagenomic and Microbiome Studies
- Nanoliter Scale High-Throughput Protein Crystallography Screening with the Echo Liquid Handler
- Optimizing EV Analysis with a CytoFLEX nano flow cytometer and FCMPASS
- Preparation of Mouse Plasma Microsamples for LC-MS/MS Analysis Using the Echo Liquid Handler
- Robust and High-Throughput SARS-CoV-2 Viral RNA Detection, Research, and Sequencing Using RNAdvance Viral and the OT-2 Platform
- Investigating the murine hepatic immune composition in diet-induced obesity using OMIP-104: Transferring an existing OMIP panel onto the CytoFLEX mosaic Spectral Detection Module
- The Valita Aggregation Pure assay: A rapid and accurate alternative for aggregation quantification of purified monoclonal antibodies
- Accurate enumeration of phytoplankton using FCM
- Accurately measures fine bubble size and particle count
- Achieving Compliant Batch Release – Sterile Parenteral Quality Control
- Adaptive Laboratory Evolution of Pseudomonas putida in the RoboLector
- Adjustment of the pH control settings in the BioLector Pro microbioreactor
- Aerobic cultivation of high-oxygen-demanding microorganisms in the BioLector XT microbioreactor
- An Analytical Revolution: Introducing the Next Generation Optima AUC
- BioLector XT 新一代高通量微型生物反应器益生菌厌氧培养工艺研究
- 使用 Multisizer 4e 库尔特颗粒计数及粒度分析仪监测贻贝/软体动物的繁殖
- Assay Assembly for Miniaturized Quantitative PCR in a 384-well Format Using the Echo Liquid Handler
- Automated 3D Cell Culture and Screening by Imaging and Flow Cytometry
- Automating a Linear Density Gradient for Purification of a Protein:Ligand Complex
- 自动化生物制药质量控制以降低成本并提高数据完整性
- Automating Bradford Assays
- Automating Cell-Based Processes
- Automating Cell Line Development
- Leveraging the Vi-CELL MetaFLEX for Monitoring Cell Metabolic Activity
- Automation of CyQuant LDH Cytotoxicity Assay using Biomek i7 Hybrid Automated Workstation to Monitor Cell Health
- Leveraging the Vi-CELL MetaFLEX for Monitoring Cell Metabolic Activity
- Automation of protein A ELISA Assays using Biomek i7 hybrid workstation
- The new Avanti J-15 Centrifuge Improves Sample Protection and Maximizes Sample Recovery
- The New Avanti J-15 Centrifuge Time Saving Deceleration Profile Improves Workflow Efficiency
- Avanti JXN Protein Purification Workflow
- Avoid the Pitfalls When Automating Cell Viability Counting for Biopharmaceutical Quality Control
- Basics of Multicolor Flow Cytometry
- Beer, Evaluation of Final Product and Filtration Efficiency
- Monitoring E. coli Cultures with the BioLector and Multisizer 4e Instruments
- Biomek基因组样品制备自动化解决方案加速研究进程
- Biomek i-Series Automated AmpliSeq for Illumina® Library Prep Kit
- Biomek i-Series Automated Beckman Coulter Agencourt RNAdvance Blood Kit
- Biomek i-Series Automated Beckman Coulter Agencourt RNAdvance Cell
- Biomek i-Series Automated Beckman Coulter Agencourt SPRIselect for DNA Size Selection
- Biomek i-Series Automated IDT® xGen Hybridization Capture of DNA libraries on Biomek i7 Hybrid Genomics Workstation
- Biomek i-Series Automated Illumina Nextera DNA Flex Library Prep Kit
- Biomek i-Series Automated Illumina® Nextera XT DNA Library Prep Kit
- Biomek i-Series Automated Illumina TruSeq DNA PCR-Free Library Prep Kit
- Biomek i-Series Automated Illumina TruSeq® Nano DNA Library Prep Kit
- Biomek i-Series Automated Illumina TruSeq® Stranded mRNA Sample Preparation Kit Protocol
- Biomek i-Series Automated Illumina TruSeq® Stranded Total RNA Sample Preparation Kit Protocol
- Biomek i–Series Automated Illumina® TruSight Tumor 170 32 Sample Method
- Biomek i-Series Automated KAPA HyperPrep and HyperPlus Workflows
- Biomek i-Series Automated New England Biolabs NEBNext® Ultra II DNA Library Prep Kit
- Biomek i-Series Automated SurePlex PCR and VeriSeq PGS Library Prep for Illumina
- Biomek i-Series Automation of the DNAdvance Genomic DNA Isolation Kit
- Cell Counting Performance of Vi–Cell BLU Cell Viability Analyzer
- Cell Line Development – Data Handling
- Cell Line Development – Limiting Dilution
- Cell Line Development – Selection and Enrichment
- 库尔特原理分析细胞
- Changes to GMP Force Cleanroom Re-Classifications
- Characterizing Insulin as a Biopharmaceutical Using Analytical Ultracentrifugation
- 洁净室常规环境监测 —— FDA 关于 21 CFR Part 11 数据完整性要求
- Fda guidance 21 cfr compliance guide for met one 3400 plus
- Cluster Count Analysis and Sample Preparation Considerations for the Vi-CELL BLU Cell Viability Analyzer
- Comparing Data Quality & Optical Resolution of the Next Generation Optima AUC to the Proven ProteomeLab on a Model Protein System
- 使用MET ONE 3400+ 进行 ISO 14644-3 洁净室自净时间测试
- Considerations of Cell Counting Analysis when using Different Types of Cells
- Consistent Cell Maintenance and Plating through Automation
- Control of Spheroid Size and Support for Productization
- Control Standards and Method Recommendations for the LS 13 320 XR
- Data-integrity-and-met-one-3400-plus-function-for-pharma
- Cydem VT Automated Clone Screening System – Generating an Antibody Standard Curve
- Cydem VT System: A Comparison to Traditional Clone Screening Platforms
- Cydem VT System Analytical Capabilities and Repeatability
- Determination of kLa values on the Cydem VT Automated Clone Screening System
- Optimize Clone Screening: Time Savings with the Cydem VT System in Monoclonal Antibody-Producing Cell Line Development Workflows
- Protein Titer Capabilities - A Comparison of the Cydem VT System to Current Technology across Various CHO Media
- Vi-CELL BLU Analyzer Data Exports and Offline Analysis Instructions
- Use Machine Learning Algorithms to Explore the Potential of Your High Dimensional Flow Cytometry Data Example of a 20–color Panel on CytoFLEX LX
- How to use R to rewrite FCS files with different number of channels
- A new approach to nanoscale flow cytometry with the CytoFLEX nano analyzer
- CytoFLEX nano 纳米流式分析仪:纳米级流式细胞仪的前沿新技术
- Detecting Moisture in Hydraulic Fluid, Oil and Fuels
- Detection of Coarse Particles in Silica Causing Cracks in Semiconductor Encapsulants
- Detection of foreign matter in plating solution using Multisizer4e
- Determination of drug-resistant bacteria using Coulter counters
- Determination of Size and Concentration of Particles in Oils
- DO-controlled fed-batch cultivation in the RoboLector®
- dsDNA Quantification with the Echo 525 Liquid Handler for Miniaturized Reaction Volumes, Reduced Sample Input, and Cost Savings
- Screening of yeast-based nutrients for E. coli-based recombinant protein production using the RoboLector Platform
- E. coli fed-batch cultivation using the BioLector® Pro
- Effective Miniaturization of Illumina Nextera XT Library Prep for Multiplexed Whole Genome Sequencing and Microbiome Applications
- Efficient clone screening with increased process control and integrated cell health and titer measurements with the Cydem VT Automated Clone Screening System
- Efficient Factorial Optimization of Transfection Conditions
- Enhancing Vaccine Development and Production
- Enumeration And Size Distribution Of Yeast Cells In The Brewing Industry
- European Pharmacopoeia EP 2.2.44 and Total Organic Carbon
- Evaluation of Instrument to Instrument Performance of the Vi-CELL BLU Cell Viability Analyzer
- Exosome-Depleted FBS Using Beckman Coulter Centrifugation: The cost-effective, Consistent choice
- Filling MicroClime Environmental Lids
- Flexible ELISA automation with the Biomek i5 Workstation
- Friction Reduction System High Performance
- Fully Automated Peptide Desalting for Liquid Chromatography–Tandem Mass Spectrometry Analysis Using Beckman Coulter Biomek i7 Hybrid Workstation
- Leveraging the Vi-CELL MetaFLEX for Monitoring Cell Metabolic Activity
- Get Control in GMP Environments
- Getting Started with Kaluza: Data Scaling and Compensation Adjustment
- Getting Started with Kaluza: Parameters
- g-Max: Added Capabilities to Beckman Coulter's versatile Ultracentrifuge Line
- A method of grading nanoparticles using ultracentrifugation in order to determine the accurate particle diameter
- HIAC Industrial – Our overview solution for fluid power testing for all applications
- High throughput cultivation of the cellulolytic fungus Trichoderma reesei in the BioLector®
- High-Throughput qPCR and RT-qPCR Workflows Enabled by Echo Acoustic Liquid Handling and NEB Luna Reagents
- A Highly Consistent BCA Assay on Biomek i-Series
- A Highly Consistent Lowry Method on Biomek i-Series
- Highly Reproducible Automated Proteomics Sample Preparation on Biomek i-Series
- High-throughput IgG quantitation platform for clone screening during drug discovery and development
- High-throughput Miniaturization of Cytochrome P450 Time-dependent Inhibition Screening Using the Echo 525 Liquid Handler
- Cell Line Development – Hit Picking
- Host Cell Residual DNA Testing in Reduced Volume qPCR Reactions Using Acoustic Liquid Handling
- How Violet Side Scatter Enables Nanoparticle Detection
- Automating the Cell Line Development Workflow
- ICH Q2 – the Challenge of Measuring Total Organic Carbon in Modern Pharmaceutical Water Systems
- ICH Q2 – The Challenge of Measuring Total Organic Carbon in Modern Pharmaceutical Water Systems
- ICH Q2 – the Challenge of Measuring Total Organic Carbon in Modern Pharmaceutical Water Systems
- Illumina Nextera Flex for Enrichment on the Biomek i7 Hybrid Genomics Workstation
- Importance of TOC measurement in WFI in light of European Pharmacopoeia change
- Improved data quality of plate-based IgG quantification using Spark®’s enhanced optics
- Increased throughput for IgG quantification using Valita Titer 384-well plates
- Integration of the Vi-CELL BLU Cell Viability Analyzer into the Sartorius Ambr® 250 High Throughput for automated determination of cell concentration and viability
- Temperature dependence of hydrodynamic radius of an intrinsically disordered protein measured in the Optima AUC analytical ultracentrifuge.
- Introducing the Cydem VT System: A high-throughput platform for fast and reliable clone screening in CLD
- Issues with Testing Jet Fuels for Contamination
- Jurkat Cell Analyses Using the Vi-CELL BLU Cell Viability Analyzer
- Leveraging the Vi-CELL MetaFLEX for Monitoring Cell Metabolic Activity
- Linearity of BSA Using Absorbance & Interference Optics
- Long Life Lasers
- LS 13 320 XR: Sample Preparation - How to measure success
- Beckman’s LS 13 320 XR Vs. Malvern Mastersizer
- Using Machine Learning Algorithms to Provide Deep Insights into Cellular Subset Composition
- Matching Cell Counts between Vi–CELL XR and Vi–CELL BLU
- 利用RoboLector提高谷氨酸棒状杆菌蛋白质产量的培养基优化研究
- MET ONE 3400+ LDAP & Active Directory connection Guide
- Method for Determining Cell Type Parameter Adjustment to Match Legacy Vi CELL XR
- 将 CytoFLEX S 流式细胞仪上设计的面板迁移至 CytoFLEX SRTl流式分选仪
- Miniaturization of an Epigenetic AlphaLISA Assay with the Echo Liquid Handler and the BMG LABTECH PHERAstar FS
- Miniaturization and Rapid Processing of TXTL Reactions Using Acoustic Liquid Handling
- Miniaturized Enzo Life Sciences HDAC1 Fluor de Lys Assays Using an Echo Liquid Handler Integrated in an Access Laboratory Workstation
- Miniaturized Enzymatic Assays with Glycerol
- Miniaturized EPIgeneous HTRF Assays Using the Echo Liquid Handler
- Miniaturized Gene Expression in as Little as 250 nL
- Miniaturized Genotyping Reactions Using the Echo Liquid Handler
- Miniaturized Multi-Piece DNA Assembly Using the Echo 525 Liquid Handler
- Miniaturized Sequencing Workflows for Microbiome and Metagenomic Studies
- Minimizing process variability in the manufacturing of bottled drinking water
- CytoFLEX SRT 上的混合模式分选
- Mode of operation of optical sensors for dissolved oxygen and pH value
- Modular DNA Assembly of PIK3CA Using Acoustic Liquid Transfer in Nanoliter Volumes
- Multi-Wavelength Analytical Ultracentrifugation of Human Serum Albumin complexed with Porphyrin
- Nanoliter Scale DNA Assembly Utilizing the NEBuilder HiFi Cloning Kit with the Echo 525 Liquid Handler
- Nanoscale Sorting with the CytoFLEX SRT Cell Sorter
- What to do now that ACFTD is discontinued
- Low-pH profiling in µL-scale to optimize protein production in H. polymorpha using the BioLector
- Optimized NGS Library Preparation with Acoustic Liquid Handling
- Optimizing Workflow Efficiency of Cleanroom Routine Environmental Monitoring
- Unveiling the Hidden Signals: Overcoming Autofluorescence in Spectral Flow Cytometry Analysis
- Spectral Flow Cytometry: A Detailed Scientific Overview
- Particle Counting in Mining Applications
- Performance of the Valita Aggregation Pure assay vs HPLC-SEC
- BioLector XT微型生物反应器小球藻光营养培养
- Astrios和CytoFLEX SRT流式分选仪的孔板分选速度比较
- Precision measurement of adipocyte size with Multisizer4e
- Principles of Continuous Flow Centrifugation
- Flow Cytometric Approach to Probiotic Cell Counting and Analysis
- Protocols for use of SuperNova v428 conjugated antibodies in a variety of flow cytometry applications
- Purifying High Quality Exosomes using Ultracentrifugation
- Purifying viral vector with VTi 90 rotor and CsCl DGUC
- JP SDBS Validation
- USP System Suitability
- Calibrating the QbD1200+ TOC Analyzer
- Quality Control of Anti-Blocking Powder Particle Size
- 符合《联邦法规 21 章》第 11 部分规定的质量控制电子记录
- Using the Coulter Principle to Quantify Particles in an Electrolytic Solution for Copper Acid Plating
- A Rapid Flow Cytometry Data Analysis Workflow Using Machine Learning- Assisted Analysis to Facilitate Identifying Treatment- Induced Changes
- Rapid Measurement of IgG Using Fluorescence Polarization
- Rapid Rabbit IgG Quantification using the Valita Titer Assay
- Leveraging the Vi-CELL MetaFLEX for Monitoring Cell Metabolic Activity
- Root Cause Investigations for Pharmaceutical Water Systems
- Screening yeast extract to improve biomass production in acetic acid bacteria starter culture
- 用CytoFLEX SRT细胞分选仪进行单细胞分选
- Leveraging the Vi-CELL MetaFLEX for Monitoring Cell Metabolic Activity
- 用CytoFLEX SRT分选稀有E-SLAM造血干细胞及其后续培养
- Unlocking Insights: The Vital Role of Unmixing Algorithms in Spectral Flow Cytometry
- Specification Comparison of Vi–CELL XR and Vi–CELL BLU
- Specifying Non-Viable Particle Monitoring for Aseptic Processing
- A Standardized, Automated Approach For Exosome Isolation And Characterization Using Beckman Coulter Instrumentation
- Streamlined Synthetic Biology with Acoustic Liquid Handling
- Switching from Oil Testing to Water and back using the HIAC 8011+ and HIAC PODS+
- SWOFF The unrecognized yet indispensable sibling of FMO
- 使用基于13色管的DURAClone干粉试剂在CytoFLEX流式细胞仪上进行人T细胞亚群的高级分析
- The scattered light signal: Calibration of biomass
- Comparative Performance Analysis of CHO and HEK Cells Using Vi-CELL BLU Analyzer and Roche Cedex® HiRes Analyzer
- Using k-Factor to Compare Rotor Efficiency
- Utilization of the MicroClime Environmental Lid to Reduce Edge Effects in a Cell-based Proliferation Assay
- Validation of On-line Total Organic Carbon Analysers for Release Testing Using ICH Q2
- Vaporized Hydrogen Peroxide Decontamination of Vi–CELL BLU Instrument
- Vertical Rotor Case Study with Adenovirus
- 采用 CytoFLEX 进行囊泡流式细胞术检测
- Automating the Valita Aggregation Pure Assay on a Biomek i-Series Liquid Handler
- Automating the Valita Titer IgG Quantification Assay on a Biomek i-Series Liquid Handling System
- Evaluating Clone Performance and Cell-Specific Productivity: Comparing the Cydem VT System and 10 L Bioreactor Cultivations
- Rapid, Automated Purification of Adeno-Associated Virus using the OptiMATE Gradient Maker
- Reducing Variability and Hands-On time in Viral Vector purification using the OptiMATE Gradient Maker
- Variability Analysis of the Vi-CELL BLU Cell Viability Analyzer against 3 Automated Cell Counting Devices and the Manual Method
- Vi-CELL BLU FAST Mode Option
- Vi-CELL BLU 符合 21 CFRPart 11的法规要求
- Viral Vector Purification with Ultracentrifugation
- 分析型超速离心技术(AUC)在脂质纳米颗粒(LNP)表征中的应用综述
- Leveraging Analytical Ultracentrifugation for Comprehensive Characterization of Lipid Nanoparticles in Drug Delivery Systems
- Whole Genome Sequencing of Microbial Communities for Scaling Microbiome and Metagenomic Studies Using the Echo 525 Liquid Handler and CosmosID
- 产品目录
- 实验步骤
- 彩页
-
案例分析
- Achieving Increased Efficiency and Accuracy in Clinical Testing
- Adenoviral Vectors Preparation
- Algae Biofuel Production
- Antibody and Media Development
- Autophagy
- B Cell Research
- Basic Research on Reproductive Biology
- Cardiovascular Disease Research
- Cell Marker Analysis
- Choosing a Tabletop Centrifuge
- Collagen Disease Treatment
- Controlling Immune Response
- Creating Therapeutic Agents
- DNA Extraction from FFPE Tissue
- English Safety Seminar
- Equipment Management
- Exosome Purification Separation
- Fast, Cost-Effective and High-Throughput Solutions for DNA Assembly
- Future of Fishing Immune Research
- Hematopoietic Tumor Cells
- High-throughput next-generation DNA sequencing of SARS-CoV-2 enabled by the Echo 525 Liquid Handler
- Hiroshima Genbaku HP Hematopoietic Tumor Testing
- iPS Cell Research
- Leveraging acoustic and tip-based liquid handling to increase throughput of SARS-CoV-2 genome sequencing
- Membrane Protein Purification X Ray Crystallography
- Organelles Simple Fractionation
- Particle Interaction
- Quality evaluation of gene therapy vector
- Retinal Cell Regeneration
- Sedimentary Geology
- Severe Liver Disease Treatment
- Tierra Biosciences reveals major molecular discovery
- Treating Cirrhosis
- University Equipment Management
- Fundamentals of Ultracentrifugal Virus Purification
- DxFLEX that provides On-site Service Support
- Improving Efficiency in Clinical FCM Workflow
- Looking to the Future of Research Support
- Opening New Possibilities for Extracellular Protein Degradation
- The Importance of FCM education and CytoFLEX
- Nanoflowcytometry for EV research
- Measuring the number of CD34 using AQUIOS
- 单页
-
专家访谈
- Background and Current Status of the Introduction of Flow Cytometers
- Bacteriological-measurements-of-soil-bacteria-in-paddy-fields
- Benefits-of-the-coulter-principle-in-the-manufacturing-for-ips-cell-derived-natural-killer-cells
- Central Diagnosis in the Treatment of Childhood Leukemia 1
- Central Diagnosis in the Treatment of Childhood Leukemia 2
- Challenges-in-viability-cell-counting
- Contribution of Cytobank to 1-cell analysis of the cancer microenvironment
- Development of technology for social implementation of synthetic biology
- Flow Cytometry Testing in Hospital Laboratories
- Fundamentals of Ultracentrifugal Virus Purification
- Tumor Suppressor Gene p53 research and DNA Cleanup Process
- Fundamentals of Ultracentrifugal Virus Purification
- Dr Yabui UCF Lecture
- Importance of Cell Cluster Volume Measurement in Regenerative Medicine
-
主题报告
- Applications of Ultracentrifugation in Purification and Characterization of Biomolecules
- Automating Genomic DNA Extraction from Whole Blood and Serum with GenFind V3 on the Biomek i7 Hybrid Genomic Workstation
- ABRF 2019: Automated Genomic DNA Extraction from Large Volume Whole Blood
- Automated library preparation for the MCI Advantage Cancer Panel at Miami Cancer Institute utilizing the Beckman Coulter Biomek i5 Span-8 NGS Workstation
- Automating Cell Line Development for Biologics
- Cell-Line Engeneering
- Characterizing the Light-Scatter Sensitivity of the CytoFLEX Flow Cytometer
- AACR 2019: Isolation and Separation of DNA and RNA from a Single Tissue or Cell Culture Sample
- Mastering Cell Counting
- Preparing a CytoFLEX for Nanoscale Flow Cytometry
- A Prototype CytoFLEX for High-Sensitivity, Multiparametric Nanoparticle Analysis
- ABRF 2019: Simultaneous DNA and RNA Extraction from Formalin-Fixed Paraffin Embedded (FFPE) Tissue
- Quantification of AAV Capsid Loading Fractions: A Comparative Study
- Using Standardized Dry Antibody Panels for Flow Cytometry in Response to SARS-CoV2 Infection
- 产品说明书
-
白皮书
- Centrifugation is a complete workflow solution for protein purification and protein aggregation quantification
- AUC Insights - Analysis of Protein-Protein-Interactions by Analytical Ultracentrifugation
- A General Guide to Lipid Nanoparticles
- Analytical Ultracentrifugation: A Versatile and Valuable Technique for Macromolecular Characterization
- Addressing issues in purification and QC of Viral Vectors
- Automation Approach to Accelerate Antibody Drug Development
- Elevate Your Extracellular Vesicle (EV) Research – An Introduction to EVs
- Enhancing Molecular Studies with Multiwavelength Analytical Ultracentrifugation
- GMP Cleanrooms Classification and Routine Environmental Monitoring
- Purification of Biomolecules by DGUC
- AUC Insights - Assessing the quality of adeno-associated virus gene therapy vectors by sedimentation velocity analysis
- AUC Insights - Sample concentration in the Analytical Ultracentrifuge AUC and the relevance of AUC data for the mass of complexes, aggregation content and association constants
- Analyzing Biological Systems with Flow Cytometry
- 亚可见颗粒物检测新进展:USP <1788>的最新修订
- Changes to USP <643> Total Organic Carbon
- Characterization of RNAdvance Viral XP RNA Extraction Kit using AccuPlex™ SARS–CoV–2 Reference Material Kit
- CytoFLEX Platform Gain Independent Compensation Enables New Workflows
- CytoFLEX Platform Flow Cytometers with IR Laser Configurations: Considerations for Red Emitting Dyes
- Evaluation of the Analytical Performance of the AQUIOS CL Flow Cytometer in a Multi-Center Study
- Simultaneous Isolation and Parallel Analysis of gDNA and total RNA for Gene Therapy
- Hydraulic Particle Counter Sample Preparation
- Inactivation of COVID–19 Disease Virus SARS–CoV–2 with Beckman Coulter Viral RNA Extraction Lysis Buffers
- Tips for Cell Sorting
- Liquid Biopsy Cancer Biomarkers – Current Status, Future Directions
- MET ONE 3400+ IT Implementation Guide
- Reproducibility in Flow Cytometry
- SuperNova v428: New Bright Polymer Dye for Flow Cytometry
- SuperNova v428: New Bright Polymer Dye for Flow Cytometry
- Japan Document
-
应用手册

