Biomek i–Series Automated Illumina® TruSight Tumor 170 32 Sample Method
Qi Wei, Ph.D., Weiming Shen, MS, and Joseph Rantus (Miami Cancer Institute) Andres Medellin and Zach Smith, MS (Beckman Coulter, Inc.)Introduction
The TruSight® Tumor 170 sequencing assay is a qualitative oncology test optimized for the detection of gene variants across 170 gene targets in nucleic acids (DNA and RNA) extracted from formalin– fixed, paraffin–embedded (FFPE) cancer tissue samples. The automated Illumina® TruSight Tumor 170 workflow supports 3 to 64 libraries. A maximum of 32 RNA samples derived from FFPE or cell lines and a maximum of 32 DNA samples (either genomic DNA or DNA isolated from the same FFPE samples as the RNA) can be prepared simultaneously into Illumina libraries using two TruSight Tumor 170 kits.
In this flyer, we demonstrate the automated performance of Illumina TruSight Tumor 170 32 Sample Method on the Biomek i5 Span–8 Genomics Workstation.
In comparison to the manual pipetting, the Illumina TruSight Tumor 170 32 Sample Method automated on Biomek platform provides:
- Reduced hands–on time and increased throughput
- Reduction in potential pipetting errors
- Standardized workflow for improved results
- Quick implementation with ready–to–implement methods
- Knowledgeable support
Spotlight
Biomek i5 Span–8 Genomics Workstation System features deliver reliability and efficiency to increase user confidence and walk–away time
- Span–8 pod with 0.5–1000 μL pipetting capability using disposable tips
- Independent 360˚ rotating gripper with offset fingers
- High deck capacity with 25 positions
- Orbital Shaker and Peltiers to provide shaking in additional to heating and cooling reagents
- Spacious, open platform design to integrate on–deck and off–deck elements (e.g. thermo cyclers)
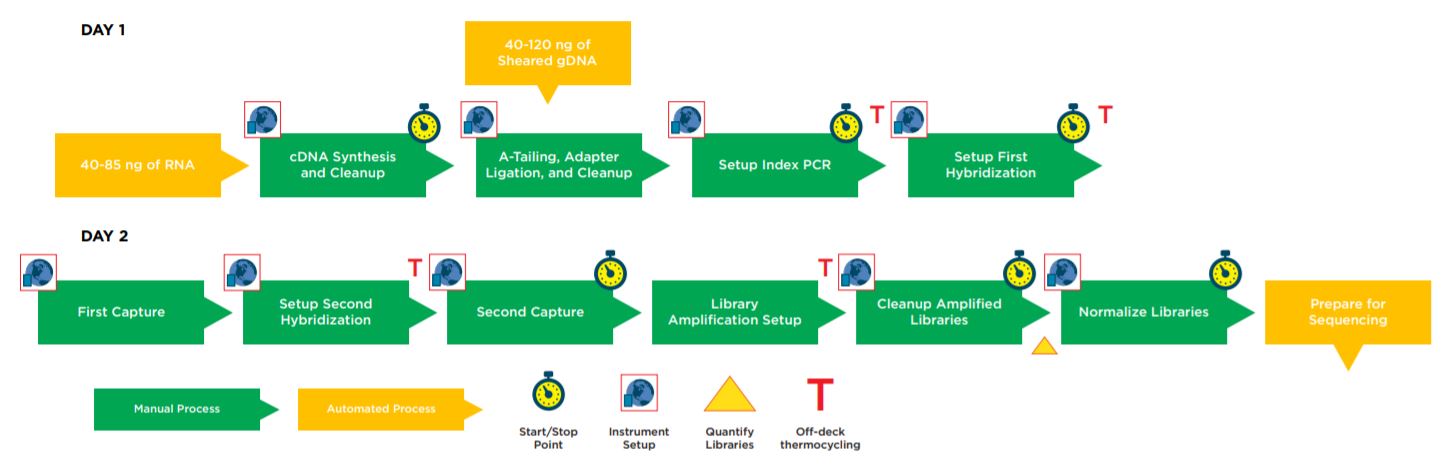
Figure 1. Illumina TruSight Tumor 170 32 Sample Method Workflow.
Automated Method
Automation provides increased efficiency, reducing the hands–on time (Table 1). The complete method can be run with full walk away capability but does include logical start and stop points assigned based on Illumina’s recommendations, providing flexibility to the users in scheduling their workflow and allowing each laboratory to address their individual requirements for sample processing and throughput.
32 DNA and 32 RNA Samples
| Day 1 | Time | |||
| Biomek Setup | Biomek Runtime | Manual procedure N | Non-Biomek Instrument Time | |
| Denature and Anneal RNA, First Strand Synthesis, Second Strand cDNA Synthesis, and cDNA Cleanup* | 15 min | 3 hr, 27 min | ||
| Covaris Shearing | 15 min | 2 hr | ||
| End Repair and A-Tailing, Ligate Adapters, and Clean | 15 min | 3 hr, 53 min | ||
| Indexing PCR | 10 min | 14 min | 45 min | |
| First Hybridization | 10 min | 38 min | Over Night | |
| Totals | 50 min | 8 hr, 12 min | 15 min | >18 hr |
| Day 2 | Time | |||
| Biomek Setup | Biomek Runtime | Manual procedure N | Non-Biomek Instrument Time | |
| First Capture | 15 min | 3 hr, 6 min | ||
| Second Hybridization | 10 min | 32 min | 1 hr, 30 min | |
| Second Capture | 15 min | 2 hr, 6 min | ||
| Amplify Enriched Libraries | 50 min | |||
| Clean Up Amplified Libraries | 15 min | 1 hr, 17 min | ||
| Library Quantification | 20 min | |||
| Normalize Libraries | 15 min | 1 hr, 32 min | ||
| Sequencing Setup | 20 min | |||
| Totals | 1 hr, 10 | 8 hr, 33 min | 40 min | 2 hr, 20 min |
*Covaris shearing of DNA samples running concurrently with Denature and Anneal RNA, First Strand Synthesis, Second Strand cDNA Synthesis, and cDNA Cleanup section
Figure 2. Illumina TruSight Tumor 170 32 Sample Method automated on the Biomek i5 Span–8 timing estimates. Timing estimates include both manual and off–deck thermocycling/incubation times
The software provides several user friendly features
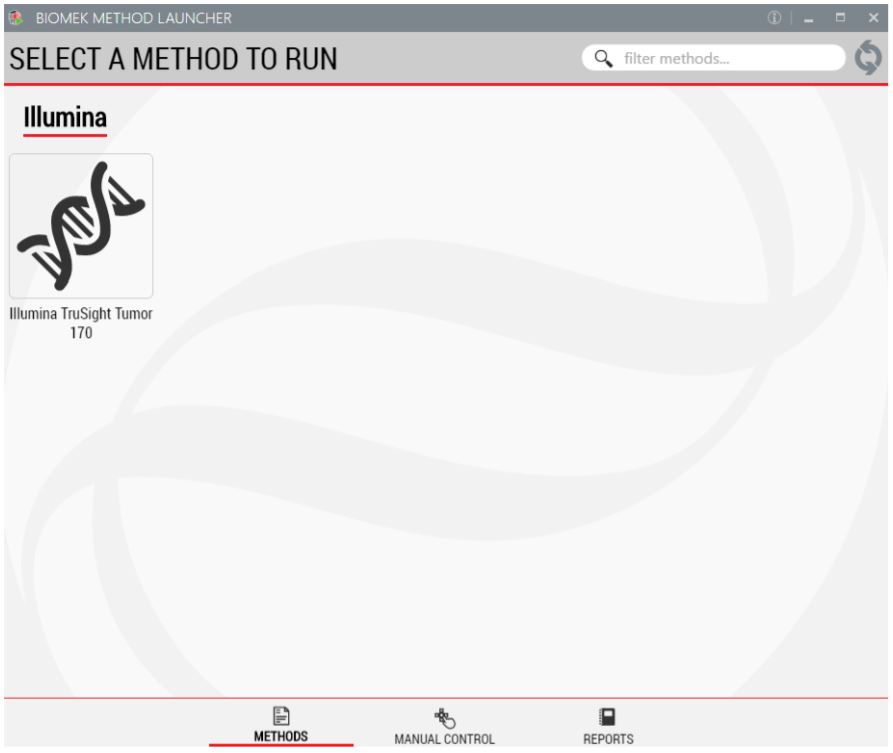
1. Biomek Method Launcher (BML)
BML is a secure interface for method implementation without affecting method integrity. It allows the users to remotely monitor the progress of the run. The manual control options provide the opportunity to interact with the instrument, if needed.
Figure 3. Biomek Method Launcher provides an easy interface to launch the method.
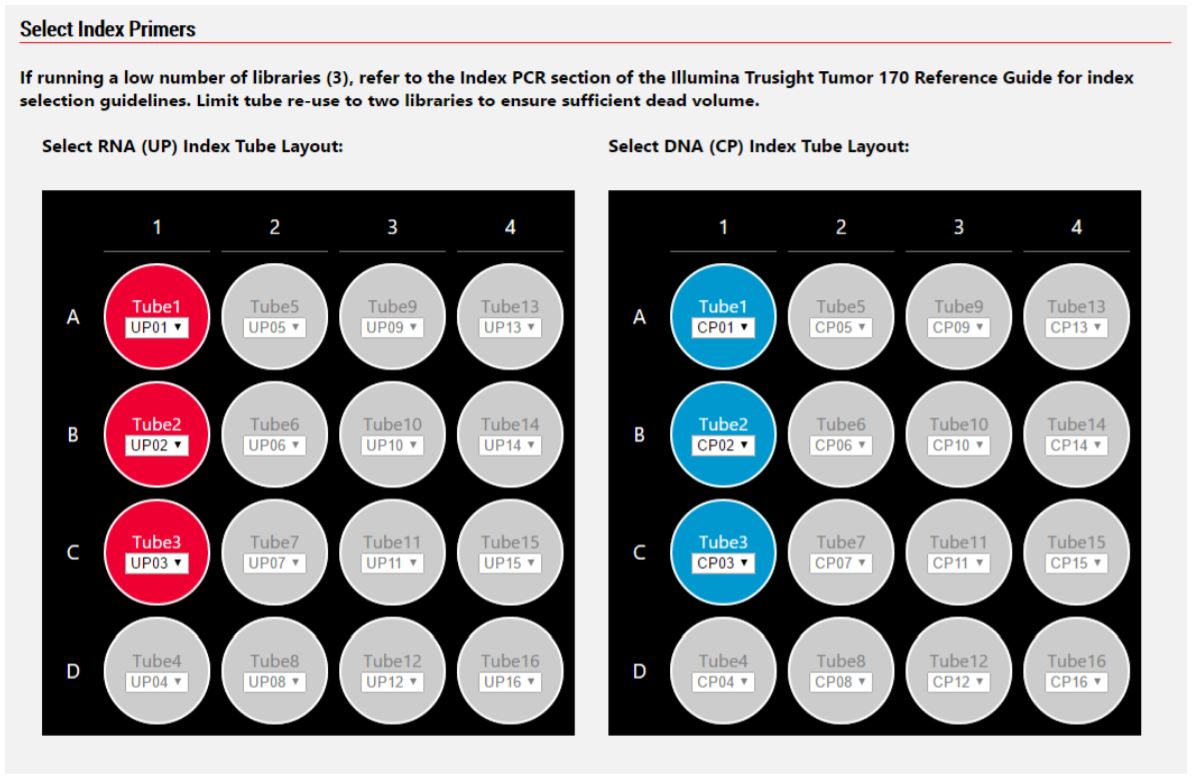
2. Method Options Selector (MOS)
MOS enables selection of plate processing and sample number options to maximize flexibility, adaptability and the ease of method execution.
Figure 4. Biomek Method Options Selector enables us to select sample number and processing options.
Figure 5. Biomek MOS allows for custom specification of CP and UP index adapters.
3. Guided Labware Setup (GLS)
GLS is generated based on options selected in the MOS, and provides the user specific graphical setup instructions with reagent volume calculation and step by step instructions to prepare reagents.
Figure 6. Guided Labware Setup indicates reagent volumes and guides the user for correct deck setup.
Experimental Design
The automated Illumina TruSight Tumor 170 32 Sample Method was installed and run at Miami Cancer Institute with a variety of samples ranging from Horizon Quantitative and Structural Reference Standards (Horizon, HD200 and HD789), Ashkenazim PGP Son Reference Standard (Horizon, GM24385), SeraSeq Fusion FFPE Reference (SeraCare, 0710–0129), and a number of in–house samples isolated with the FormaPure Total (Beckman Coulter). 108 DNA samples and 80 RNA samples were processed on the Biomek i5 Span–8 over the course of 13 runs. Following sequencing, libraries were analyzed using the TruSight Tumor 170 App on BaseSpace (basespace.illumina.com).
Results
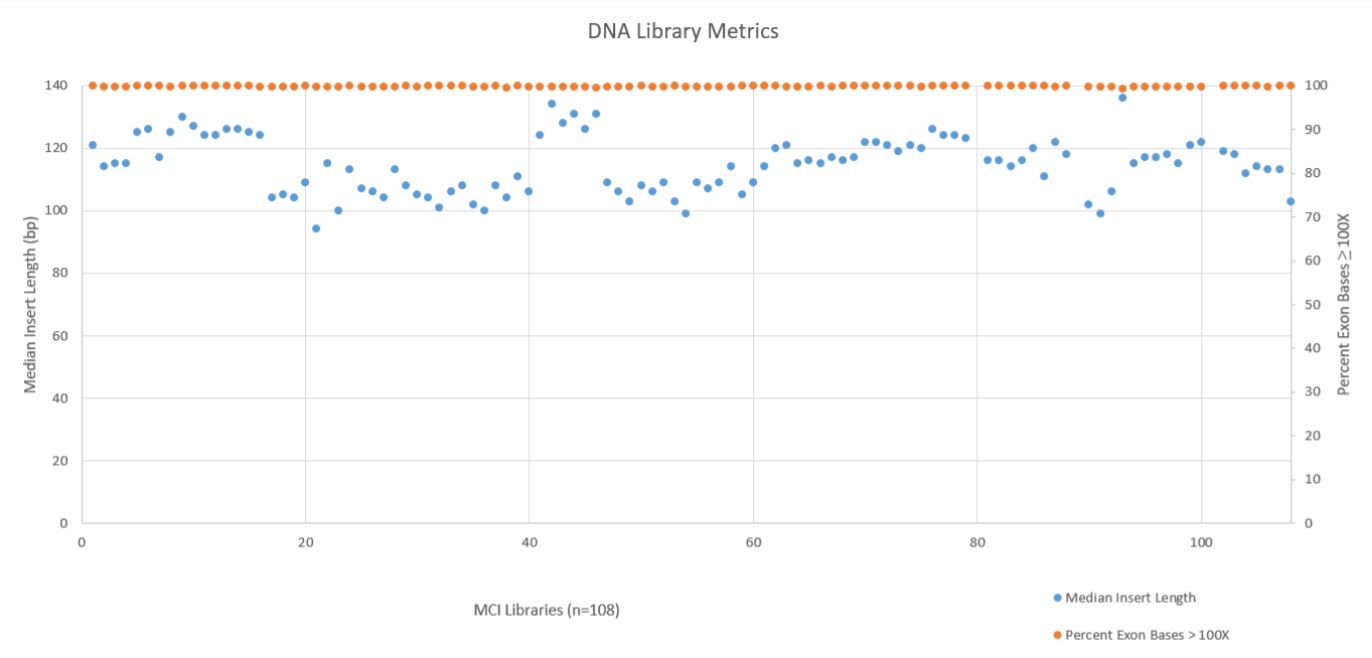
A robust and reliable automation solution for NGS library prep is essential to take full advantage of Illumina’s sequencing technology. Automation saves up valuable time and money and helps the researchers to obtain an efficient process with more walk–away time. Two DNA metrics, Median Insert Length and the Percent Exon Bases ≥ 100X Coverage, are shown below in Figure 7. These metrics show that the median insert length across all 108 libraries was reasonable consistent (mean 115bp with a standard deviation of 9bp), while the exon coverage at 100X or greater was very high across all libraries (mean 99.78% with a standard deviation of 0.09%). All libraries passed the minimum quality threshold (median insert length ≥ 79bp and Percent Exon Bases ≥ 100X greater than 95%) as defined by the TruSight Tumor 170 BaseSpace App.
Figure 7. DNA Library Metrics.
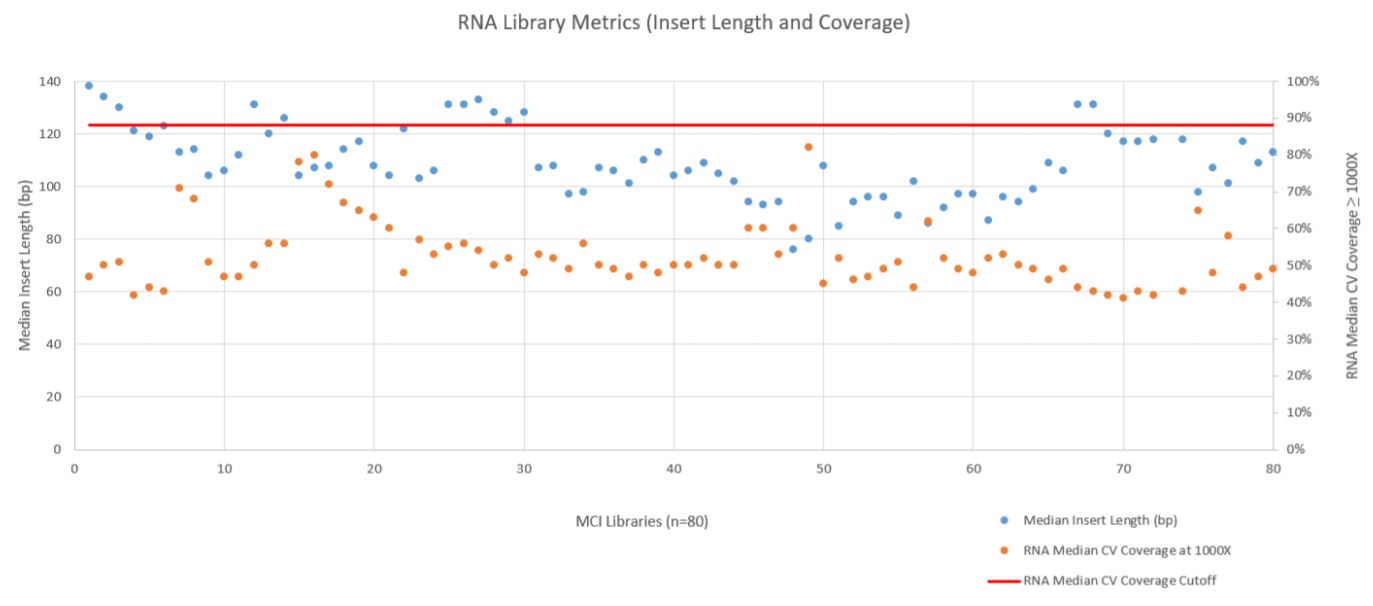
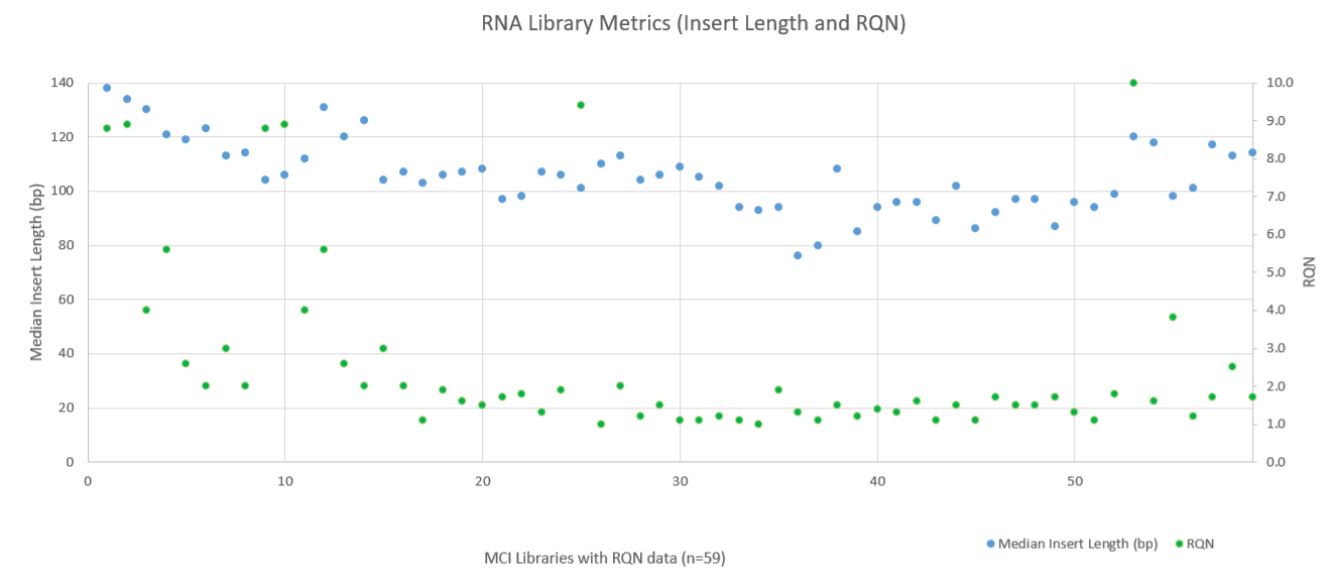
RNA sequencing metrics (presented in Figure 8), show that for the 80 samples sequenced median insert length varied somewhat more than the DNA libraries (mean 109bp with a standard deviation of 14bp). RNA Median CV Coverage at 1000X was more consistent (mean 0.53 with a standard deviation of 0.09). RNA quality (utilizing the RNA Quality Number from the Advanced Analytical Fragment Analyzer) for the samples ranged from 10 (highest) to 1 (lowest) for the 59 samples that RQN values were available for. Purchased controls (samples 1–16) show a wide range of variability in terms of RQN as related to median insert length compared to the in–house samples (samples 17–65) as shown in Figure 9. However, all libraries passed the minimum quality thresholds (Median Insert Size ≥ 63bp and RNA Median CV Coverage ≥ 1000X less than or equal to 0.88) as defined by the TruSight Tumor 170 BaseSpace App.
Figure 8. RNA Metric (Fragment Length and Coverage).
Figure 9. RNA Metrics (Fragment Length and RQN Values).
Summary
We’ve demonstrated automation of the Illumina TruSight Tumor 170 Library Preparation kit on the Biomek i5 Span-8 NGS Workstation delivers libraries that yield quality results over a variety of sample inputs.