Illumina Nextera Flex for Enrichment on the Biomek i7 Hybrid Genomics Workstation
Introduction
The Illumina Nextera Flex for Enrichment assay is a flexible target enrichment next generation sequencing (NGS) kit that works across a variety of applications ranging from fixed panels to whole exome sequencing. The kit utilizes Illumina’s On-Bead Tagmentation chemistry for library construction, which uses bead-linked transposomes to mediate the tagmentation reaction to enable more uniform results over a wide range of sample types including genomic DNA (gDNA) samples (10 ng to 1000 ng input) or DNA extracted from formalin-fixed, paraffin-embedded (FFPE) tissue samples (50 ng to 1000 ng input). Additionally, supplemental protocols allow for the use of blood or saliva as direct inputs to the Illumina Nextera Flex for Enrichment assay. The target enrichment portion of the assay uses a single 90-minute hybridization to allow for rapid capture and enrichment of the library pools. The entire manual protocol can be completed in less than 6.5 hours for 12 gDNA libraries pooled and enriched as a single 12-plex capture pool.
In this flyer, we describe and demonstrate the automated performance of the Illumina Nextera Flex for Enrichment assay on the Biomek i7 Hybrid Genomics Workstation. The automation solution can support library construction from between 1 and 96 libraries and target capture of between 1 and 96 library pools, allowing the user to select any plexity between 1 and 12. The automation solution supports both supplemental protocols for beginning library construction with blood or saliva inputs as well gDNA or FFPE DNA. During the target enrichment stage, the automation solution also supports the supplemental protocol for library pool concentration using AMPure XP on the Biomek i7 or off-instrument vacuum concentration in addition to on-deck hybridization with an integrated thermocycler. The automation solution can process 96 samples in approximately 10 hours, 30 minutes of total time with 45 minutes of hands-on time.
In comparison to the use of manual pipetting, the Illumina Nextera Flex for Enrichment automated on Biomek platform provides:
- Reduced hands-on time and increased throughput
- Reduction in potential pipetting errors
- Standardized workflow for improved results
- Quick implementation with ready-to-implement methods
- Knowledgeable support
Spotlight
Biomek i7 Dual Hybrid (Multichannel 96, Span-8) Genomics Workstation
System features deliver reliability and efficiency to increase user confidence and walk-away time
- 1200 μL Multichannel 96 head with 1 μL to 1000 μL pipetting capability
- Span-8 pod with disposable tips for performing transfers between 1 μL to 1000 μL
- Enhanced Selective Tip pipetting to transfer custom arrays of samples using the Multichannel 96 head
- Independent 360˚ rotating grippers with offset fingers
- High deck capacity with up to 45 positions
- Shaking, heating/cooling, and tip washing for controlling sample processing
- Spacious, open platform design to integrate on-deck and off-deck elements (e.g. thermocyclers)
Figure 1. Biomek i7 Genomics Workstation
Automated Method
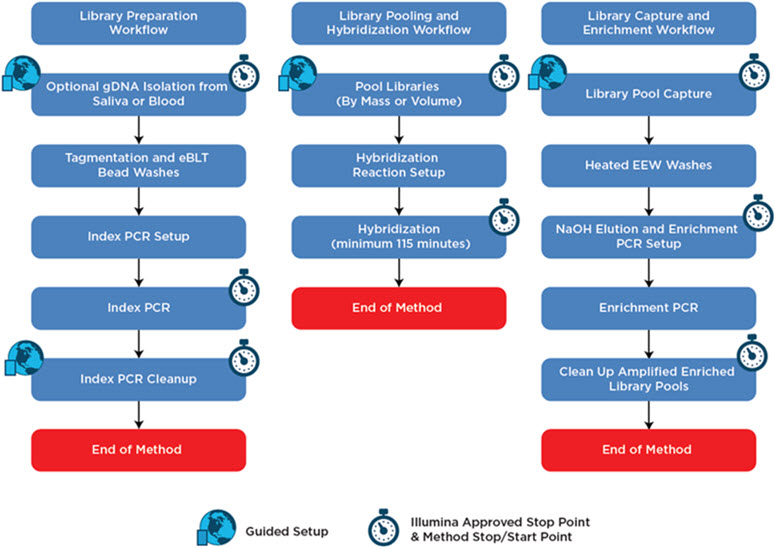
The automated Illumina Nextera Flex for Enrichment method is constructed in a modular fashion that follows the manual assay’s recommended start and stop points, allowing the operator flexibility in performing the assay and allowing the automation method to be easily deployed in pre-PCR and post-PCR laboratory spaces. Enzymatic incubations and PCR reactions can be performed on-deck with an integrated thermocycler or with an off-deck thermocycler. Target capture requires the use of the on-deck thermocycler to heat the wash buffers used for washing the captured library pools.
Figure 2. Illumina Nextera Flex for Enrichment automation method workflow.
Automation provides increased efficiency by reducing hands-on time. In the table below, we detail the estimated time to complete the three workflows of the automation method with different sample number inputs.
| Library Construction |
8 Libraries |
24 Libraries |
96 Libraries |
| Prepare Reagents/Set Up Biomek | 15 min |
15 min | 15 min |
| Method Run |
2 hr, 25 min | 2 hr, 44 min |
3 hr, 44 min |
| Total | 2 hr, 40 min |
2 hr, 59 min |
3 hr, 59 min |
| Library Pooling and Hybridization Setup |
96 Libraries to 8 12-plex Pools |
96 Libraries To 24 4-Plex Pools |
96 Libraries To 96 1-Plex Pools |
| Prepare Reagents/Set Up Biomek | 15 min |
15 min | 15 min |
| Method Run |
2 hr, 5 min | 2 hr, 13 min |
2 hr, 29 min |
| Total | 2 hr, 20 min |
2 hr, 28 min |
2 hr, 44 min |
| Library Pool Capture and Enrichment |
8 Pools |
24 Pools |
96 Pools |
| Prepare Reagents/Set Up Biomek | 15 min |
15 min | 15 min |
| Method Run |
2 hr, 49 min | 3 hr, 2 min |
3 hr, 43 min |
| Total | 3 hr, 4 min |
3 hr, 17 min |
3 hr, 58 min |
* Timing estimate assumes gDNA as a starting material and includes incubations and thermocycling. Hybridization time was estimated at minimum required time.
Table 1. Estimated run times for processing 8, 24, and 96 libraries in library construction and 8, 24, and 96 library pools in target enrichment. Timing estimates for method run times were obtained from Biomek software.
The software provides several user-friendly features such as
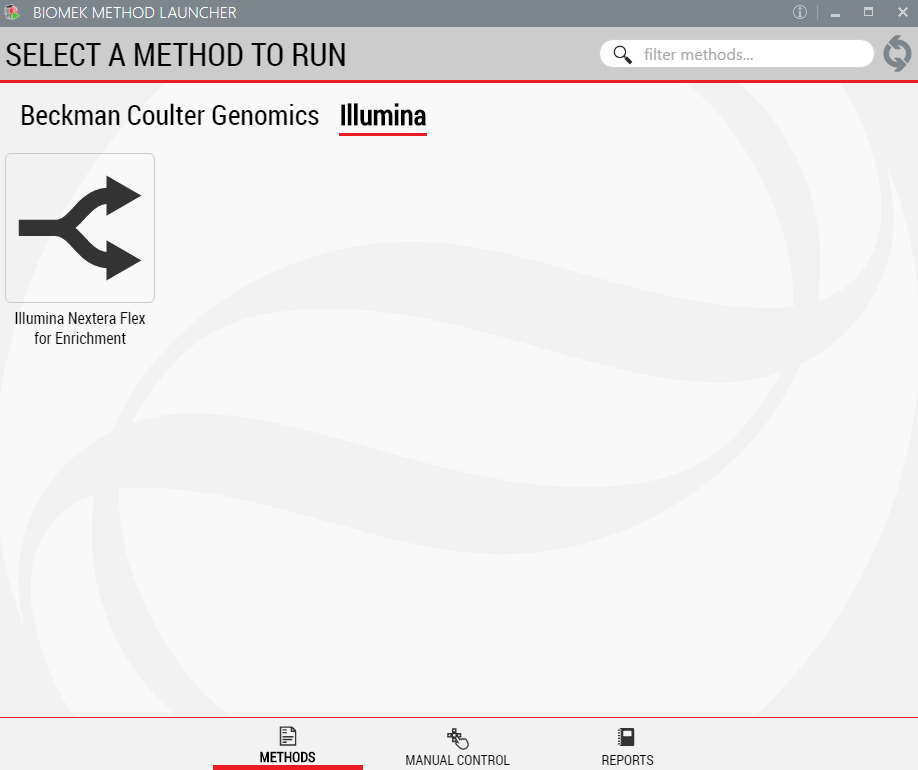
1. Biomek Method Launcher (BML)
BML is a secure interface for method implementation without affecting method integrity. It allows the users to remotely monitor the progress of the run. The manual control options provide the opportunity to interact with the instrument if needed. The automated method may also be run through the standard Biomek software if Biomek Method Launcher is unavailable.
Figure 3. Biomek Method Launcher provides an easy interface to launch the method.
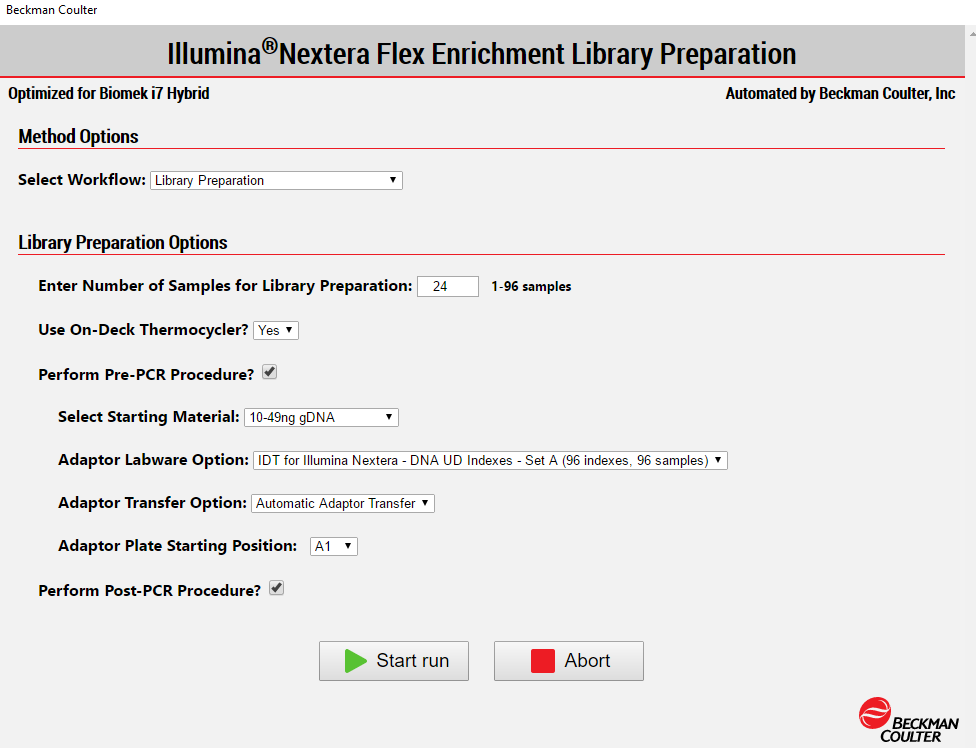
2. Method Options Selector (MOS)
MOS enables selection of plate processing and sample number options to maximize flexibility, adaptability, and the ease of method execution.
Figure 4. Biomek Method Options Selector enables user to select the desired workflow, sample number, and a variety of workflow customization options.
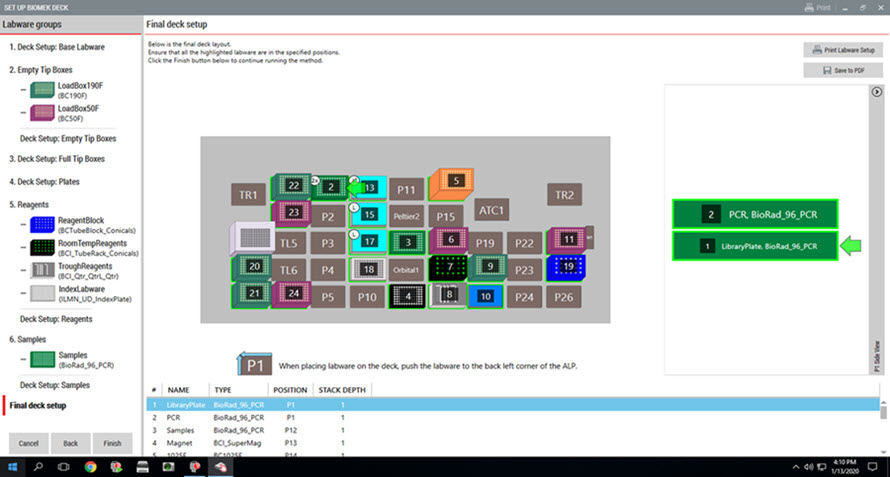
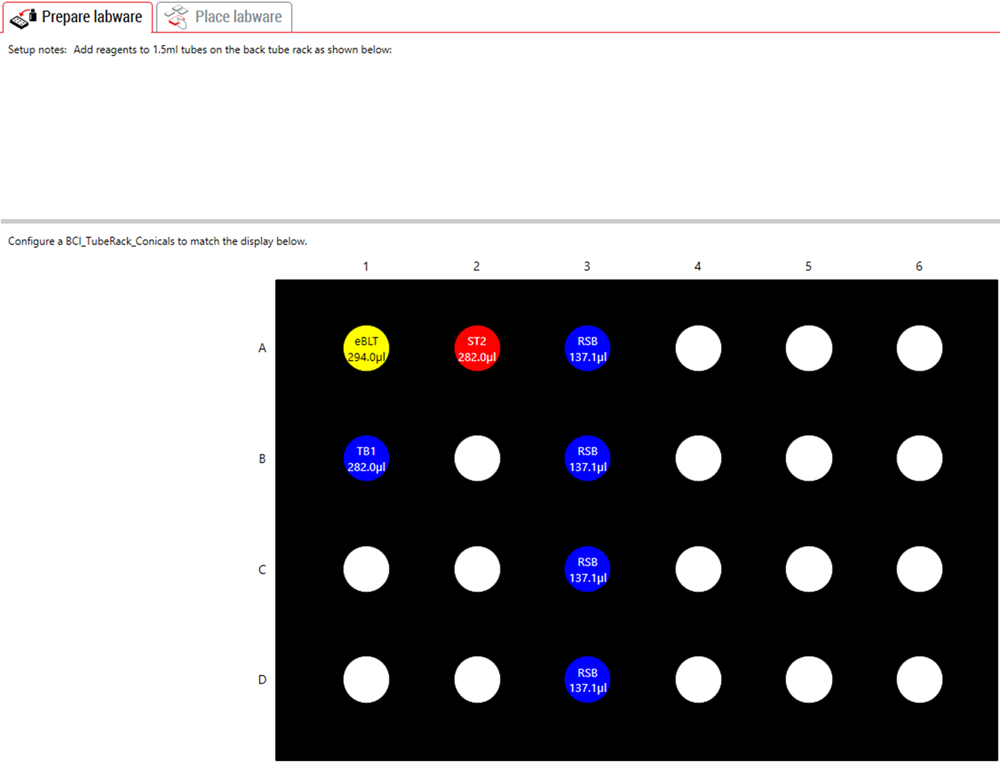
3. Guided Labware Setup (GLS)
GLS is generated based on options selected in the MOS and provides the user specific graphical setup instructions with reagent volume calculation and step by step instructions to prepare reagents. Deck Optix Final Check (part of BML) utilizes the Biomek’s built-in camera system to verify labware placement to provide another layer of protection against incorrect instrument setup.
Figure 5. Guided Labware Setup indicates reagent volumes and guides the user for correct deck setup.
Experimental Design
Coriell DNA (NA12878) was obtained from the Coriell Institute and utilized for library construction. Nextera Flex for Enrichment library construction was then performed on 48 Coriell samples (75 ng gDNA input per library). Following library construction, 48 libraries were then pooled into 4 12-plex pools by volume and ran through Nextera Flex for Enrichment target enrichment using the Illumina Coding Exome Panel (CEX). All incubations, PCR steps, and hybridizations were performed using the on-deck thermocycler. Following target enrichment, one Coriell 75ng input pool (Pool1) was sequenced on the Illumina NextSeq using a 300 cycle High Output v2 reagent kit. The Coriell Library Pool sequencing run generated 129 Gb of sequence with Q30 scores > 89% of total bases. Sequencing data was analyzed using the Illumina BaseSpace Enrichment App with default settings.
Results
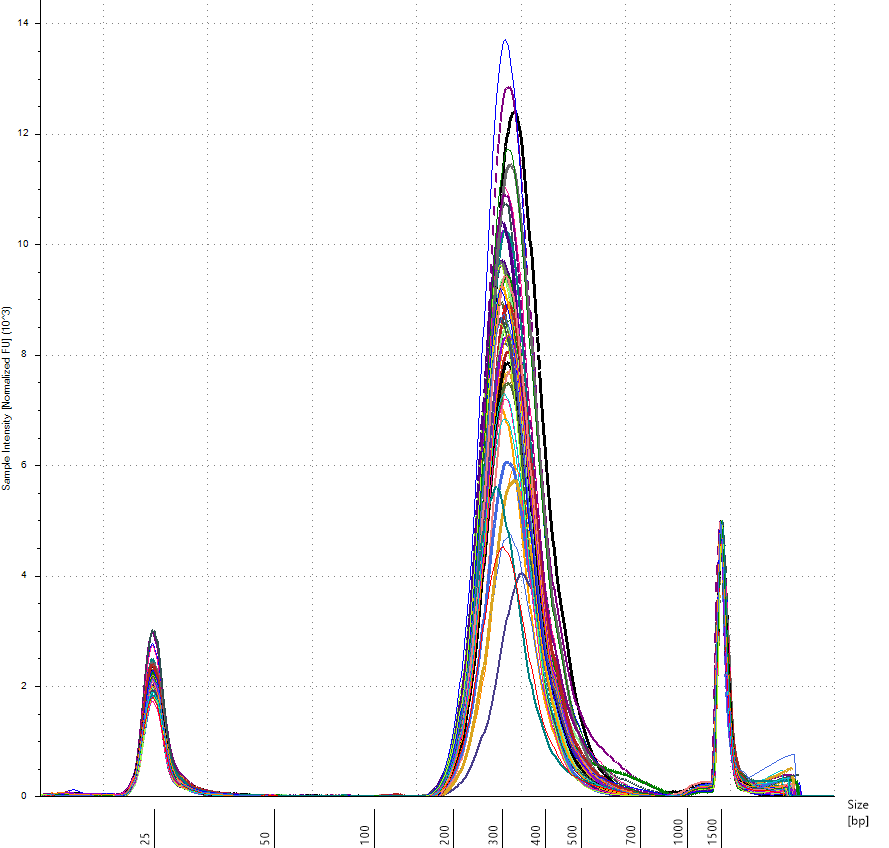
Figure 6. Nextera Flex for Enrichment libraries following library construction assayed on the Agilent 2200 TapeStation using D1000 ScreenTape.
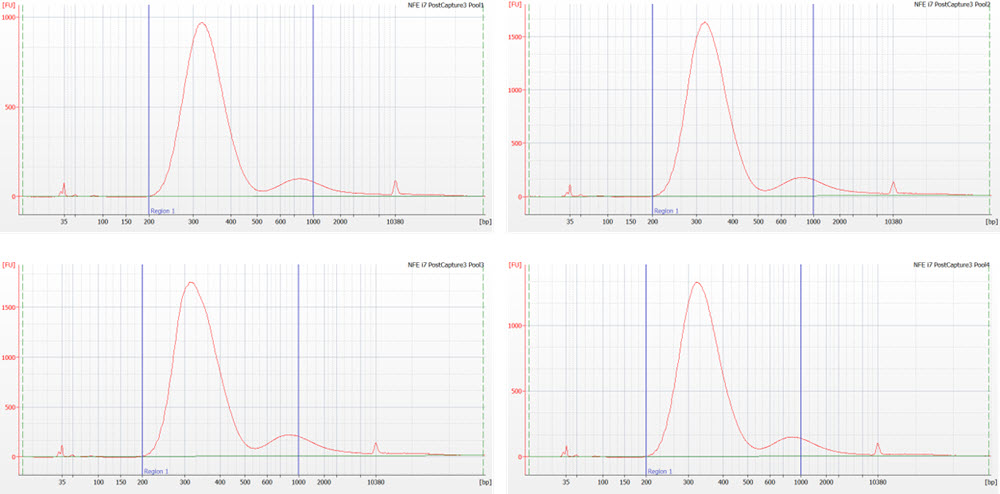
Figure 7. Nextera Flex for Enrichment library pools following target enrichment assayed on the Agilent 2100 Bioanalyzer using a DNA High Sensitivity chip.
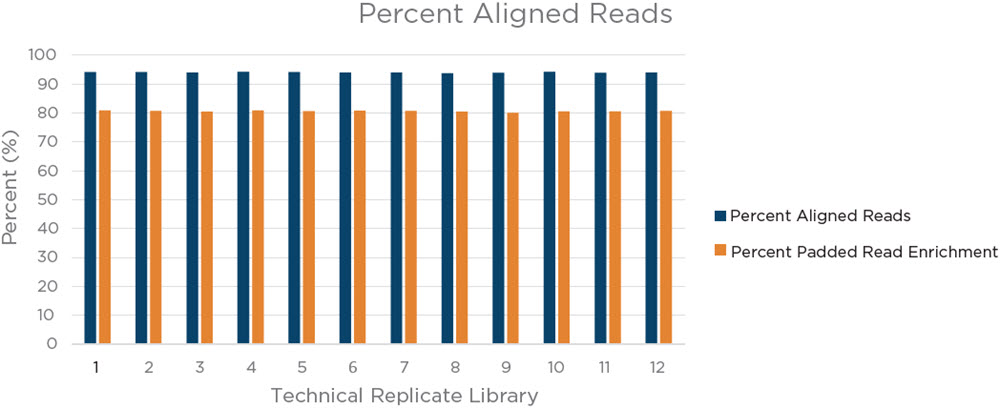
Figure 8. Coriell 75 ng input library pool percent aligned reads and percent padded read enrichment. High percent aligned reads (94% or higher in all libraries) indicates an efficient library preparation from the samples and a relative lack of non-usable reads. A high percent padded read enrichment (80% or greater for all libraries), is indicative of an efficient library capture and enrichment process, as the majority of the reads are from the regions targeted by the Illumina Coding Exome Panel.
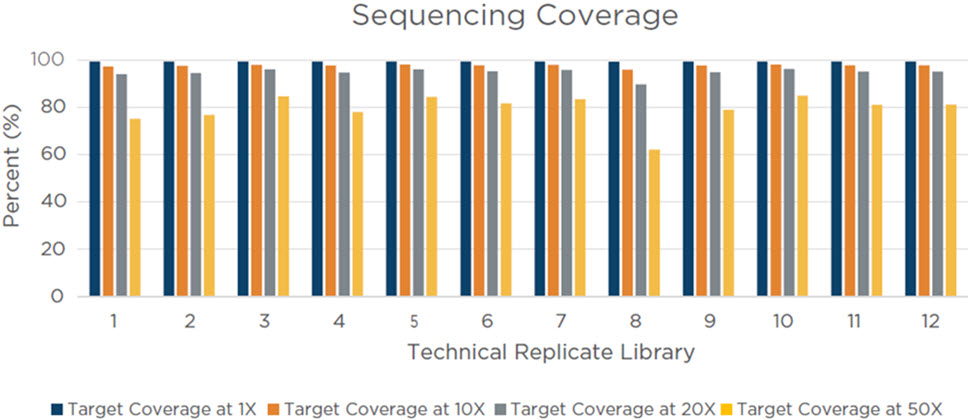
Figure 9. Coriell 75 ng input library pool target coverage. This metric measures the depth of sequencing coverage across the targeted regions of the Illumina Coding Exome Panel (n = 214,126). For each of the 12 technical replicate libraries, 80% or more of the targeted regions were covered with an average of 20 reads over each base in each targeted region, indicating an efficient hybridization and capture process.
Summary
We’ve demonstrated that automation of the Illumina Nextera Flex for Enrichment assay on the Biomek i7 Hybrid Genomics Workstation delivers libraries that yield high quality sequencing results. By providing a robust and reliable automation solution for the Illumina Nextera Flex for Enrichment assay on the Biomek i7 Hybrid Genomics Workstation, we can help researchers to take full advantage of Illumina’s sequencing technology.
The Illumina Nextera Flex for Enrichment library preparation kit is for Research Use Only. The Illumina Nextera Flex for Enrichment library preparation kit is not for use in diagnostic procedures. The Biomek Workstation is not intended or validated for use in the diagnosis of disease or other conditions. Beckman Coulter makes no warranties of any kind whatsoever, express or implied, with respect to this protocol, including but not limited to warranties of fitness for a purpose or merchantability or that the protocol is non-infringing. All warranties are expressly disclaimed. Your use of the method is solely at your own risk, without recourse to Beckman Coulter. Not intended or validated for use in the diagnosis of disease or other conditions. This protocol is for demonstration only and is not validated by Beckman Coulter.
Biomek Method Launcher software package must be purchased separately.


 <>/a
<>/a