什么是转录组学?
转录组学 专注于探究生物样品中,在 RNA 生成与降解的动态平衡中形成的 RNA 分子群体。该学科的研究目标不仅局限于 RNA 转录本的数量统计,更旨在揭示由同一转录本进行剪接(splicing)、多聚腺苷酸化(polyadenylation)、3’至5’端加工(end-processing)及其他转录后修饰过程(如microRNA 的调控作用)中 RNA 分子的多样性。
转录组学有助于分析和解释细胞在不同条件下的基因表达模式,帮助研究人员深入了解细胞的功能基因组学、调控机制和各项功能。
作为四大组学之一,转录组学对于科学家全面揭示生物系统的整体功能和调控机制做出了不可或缺的贡献。转录组学数据被广泛应用于药物开发、疾病诊断与预后评估、个性化医疗、癌症研究、神经科学、农业育种及环境监测等诸多领域。
转录组学研究面临的挑战
取得突破性发现的同时,研究人员同样面临各项挑战,包括:
- 难以从特定生物样品中有效提取 RNA
- 无法准确检测低丰度转录本
- 数据结果不完整或存在偏差
- 分析结果不一致、不可靠
- RNA 样品质量低且不稳定
- 执行多步骤实验方案时易出现人为错误
- 技术繁杂且耗时费力
- 实验成本高昂
这些挑战在很大程度上能够通过借助可信赖的液体处理系统,配合高质量试剂与整合仪器,实现 转录组学工作流程的自动化 而得以有效解决。
转录组学自动化解决方案
自 1986 年推出开创性的 Biomek 1000 自动化工作站以来,贝克曼库尔特生命科学始终致力于推动转录组学研究的持续发展。如今,我们的转录组学领域产品组合不仅因先进的 自动化移液技术而广受赞誉,更因卓越的 整合能力、用户友好的软件界面,以及 特有的 SPRI顺磁珠技术基因组试剂而声名远扬。我们持续与业界备受信赖的企业积极拓展合作关系,为用户打造全方位、无缝集成的端到端转录组学解决方案。
以下为交互式工作流程示例: 阐述我们的解决方案在常用转录组测序技术(RNA-Seq)中的应用。
我们的转录组学解决方案优势
自动化液体处理解决方案
- Biomek i-Series 自动化工作站:
Pod Volume Inaccuracy CV Span-8 0.5 μL < 0.12% < 4.58% 900 μL < 0.102% < 0.392% MC-384 0.5 μL < 0.38% < 5.06% MC-96 950 μL < 0.1% <0.28% - Echo 声波移液系统赋能用户精准转移低至 2.5 nL体积 的样品和试剂。 测定结果精准、稳定、可再现,无需重复测定 。
- Biomek i-Series 自动化工作站:
- 网格化工作台面,宽敞、开放的平台设计,多达 45 个标准板位
- 多种的移液通道配置:支持从单个灵活的 Span-8 加样器到双多通道加样器(96/384)。
- Echo 声波移液器支持任意孔间快速、精准、无接触式移液,以往 长达数小时 的 DNA/RNA 均一化、Barcode 标记和文库 Pooling 步骤,现在 仅需几分钟即可完成。
- Biomek Echo One 高通量基因组学整合解决方案,将支持反应体系微量化的声波移液技术与 Biomek i系列工作站强大而灵活的功能全面结合
- 我们的精英专家团队成功整合60 多家制造商的 300 多款第三方设备,为用户打造业内领先的实验室全自动化解决方案。
- Biomek i-Series 自动化工作站:无缝集成实验室信息管理系统(LIMS),支持连接 AI 工具和机器学习算法,实现数据高效处理。
试剂
单细胞转录组学
单细胞转录组学旨在深入探究单个细胞内部的高分辨率基因表达谱。与适用于分析细胞群体整体基因表达趋势的转录组分析有所不同,单细胞方法能够精确揭示独特的转录程序,并有效识别稀有细胞群及其特定状态。
单细胞转录组学帮助研究人员更深入地理解细胞功能、相互作用和异质性,为肿瘤学、免疫学、神经科学等领域带来深刻洞见。
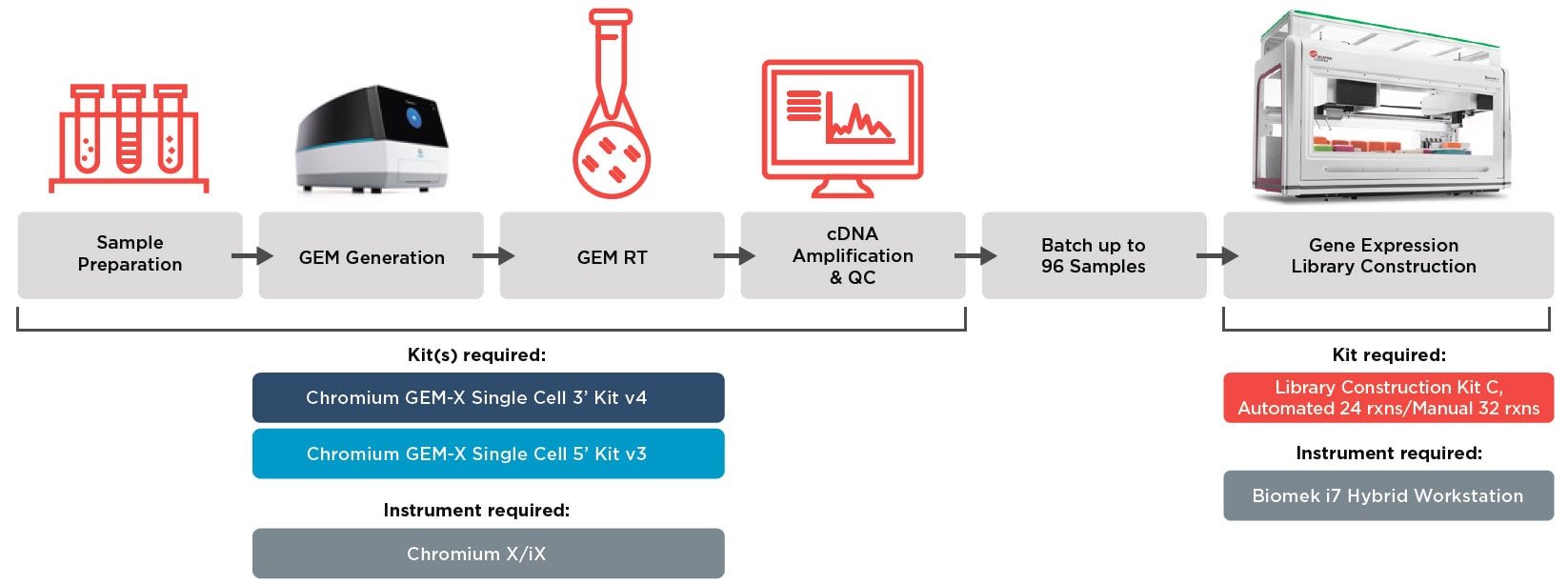
凭借先进的自动化解决方案,我们帮助 10X Genomics 公司成功简化单细胞转录组学应用 Chromium™ GEM-X 单细胞基因表达工作流程:
用户评价

Biomek i7 工作站帮助我们(Single Cell Discoveries)显著优化了服务工作流程,其强大的自动化能力不仅为我们节省了大量时间,还有效降低了人为错误风险,确保了数据的稳定性和可靠性。Biomek i7 支持我们快速、一致地处理大量样品,这种优势对于生成高质量、可重复的数据至关重要,其灵活性和多功能性让我们能够轻松定制实验方案,满足各种转录组学应用的特定需求。此外,Biomek 团队的全程贴心支持和全面培训服务,帮助我们熟练操作并充分利用该平台。
Oriol Llorà Batlle 博士,Single Cell Discoveries B.V. 研发团队负责人
癌症研究中的转录组学应用
转录组学在癌症研究 中扮演着至关重要的角色,提供癌细胞和组织内时空基因表达的深刻洞察。这些信息对于研究人员揭示驱动癌症进展、转移以及治疗耐药性的分子机制具有重要意义。
识别和了解癌症中失调的基因和通路,有助于发现新疗法的潜在靶点。通过对比分析癌组织与正常组织的转录组学特征,研究人员可精确识别特定 RNA 分子,这些分子可作为早期癌症检测、预后评估以及监测治疗反应的重要生物标志物。
癌症研究的主要挑战是癌症的复杂性和异质性。肿瘤是由多个细胞群体组成的复杂生态系统,每种细胞类型都有其独特的基因表达谱。
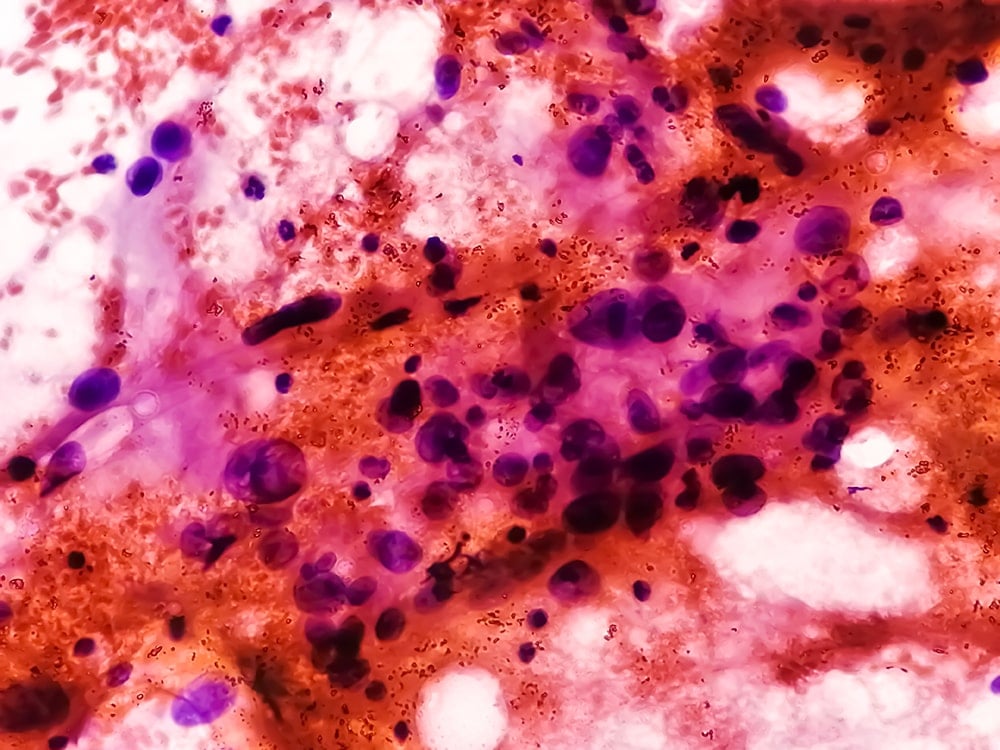
梭形细胞肉瘤细胞学
我们支持自动化的 SPRI 顺磁珠技术基因组试剂,为研究人员提供精准可靠的可重复数据,这些特性对于整体研究而言不可或缺,特别是在识别稀有癌细胞群的研究中更为关键。我们的试剂盒能够从癌症研究中最常用的样品类型之一 – 福尔马林固定石蜡包埋(FFPE)组织中高效提取核酸。此外,我们的 FormaPure XL Total 试剂盒还支持您从同一份样品中同时提取 DNA 和 RNA,无需对样品进行分割。
特色资料
Biomek i-Series Liquid Handlers
- Yao Z. et al. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature. 2021. doi: 10.1038/s41586-021-03500-8.
- Kind D. et al. Automation enables high-throughput and reproducible single-cell transcriptomics library preparation. SLAS Technol. 2022. doi: 10.1016/j.slast.2021.10.018.
- Santacruz D. et al. Automation of high-throughput mRNA-seq library preparation: a robust, hands-free and time efficient methodology. SLAS Discov. 2022. doi: 10.1016/j.slasd.2022.01.002.
Echo Acoustic Liquid Handlers
- Mayday M.Y. et al. Miniaturization and optimization of 384-well compatible RNA sequencing library preparation. PLoS ONE. 2019. doi: 10.1371/journal.pone.0206194.
- Ye C. et al. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat Commun. 2018. doi: 10.1038/s41467-018-06500-x.
- Larman H.B. et al. Sensitive, multiplex and direct quantification of RNA sequences using a modified RASL assay. Nucleic Acids Res. 2014. doi: 10.1093/nar/gku636.
- Harrill J.A. et al. High-Throughput Transcriptomics Platform for Screening Environmental Chemicals. Toxicol Sci. 2021. doi: 10.1093/toxsci/kfab009.
- Li J. et al. DRUG-seq Provides Unbiased Biological Activity Readouts for Neuroscience Drug Discovery. ACS Chem Biol. 2022. doi: 10.1021/acschembio.1c00920.
- Thomas J.R. et al. Enhancing the Small-Scale Screenable Biological Space beyond Known Chemogenomics Libraries with Gray Chemical Matter─Compounds with Novel Mechanisms from High-Throughput Screening Profiles. ACS Chem Biol. 2024. doi: 10.1021/acschembio.3c00737.
- Bush E.C. et al. PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nat Commun. 2017. doi: 10.1038/s41467-017-00136-z.
- Huot L. et al. Spodoptera frugiperda transcriptional response to infestation by Steinernema carpocapsae. Sci Rep. 2019. doi: 10.1038/s41598-019-49410-8.
- Isobe Y. et al. Manumycin polyketides act as molecular glues between UBR7 and P53. Nat Chem Biol. 2020. doi: 10.1038/s41589-020-0557-2.
Genomic Reagents
- Finseth F.R. and Harrison R.G. A comparison of next generation sequencing technologies for transcriptome assembly and utility for RNA-Seq in a non-model bird. PLoS One. 2014 . doi: 0.1371/journal.pone.0108550.
- Callari M. et al. In-depth characterization of breast cancer tumor-promoting cell transcriptome by RNA sequencing and microarrays. Oncotarget. 2016. doi: 10.18632/oncotarget.5810.
- Zhang Z. et al. Blood RNA alternative splicing events as diagnostic biomarkers for infectious disease. Cell Rep Methods. 2023. doi: 10.1016/j.crmeth.2023.100395.
- Haile S. et al. Automated high throughput nucleic acid purification from formalin-fixed paraffin-embedded tissue samples for next generation sequence analysis. PLoS One. 2017. doi: 10.1371/journal.pone.0178706.
- Gharaie S. et al. Single cell and spatial transcriptomics analysis of kidney double negative T lymphocytes in normal and ischemic mouse kidneys. Sci Rep. 2023. doi: 10.1038/s41598-023-48213-2.
- Gaisser K.D. et al. High-throughput single-cell transcriptomics of bacteria using combinatorial barcoding. Nat Protoc. 2024. doi: 10.1038/s41596-024-01007-w.
- Zou S. et al. Multi-omic profiling reveals associations between the gut microbiome, host genome and transcriptome in patients with colorectal cancer. J Transl Med. 2024. doi: 10.1186/s12967-024-04984-4.
- Alfonso-Gonzalez C. et al. Identification of regulatory links between transcription and RNA processing with long-read sequencing. STAR Protoc. 2023. doi: 10.1016/j.xpro.2023.102505.
- Pérez-Mojica J.E. et al. Continuous transcriptome analysis reveals novel patterns of early gene expression in Drosophila embryos. Cell Genom. 2023. doi: 10.1016/j.xgen.2023.100265.
我们的合作伙伴







Not intended or validated for use in the diagnosis of disease or other conditions.
ECHO is a trademark or registered trademark of Labcyte Inc. in the United States and other countries. Labcyte is a Beckman Coulter company.