RNA Isolation from Bacteria
Sequencing RNA isolated from bacterial cultures is a common method for determining what genes are being expressed in the culture. Sequencing assays usually require high quality RNA. PoP team members working with a customer optimized a protocol using the RNAdvance Tissue kit for use with bacteria by the addition of lysozyme.
Protocol:
RNA isolation from Bacteria using RNAdvance Tissue
RNA Extraction from E. coli, B. subilis and S. saliverius using RNAdvance Tissue Kit
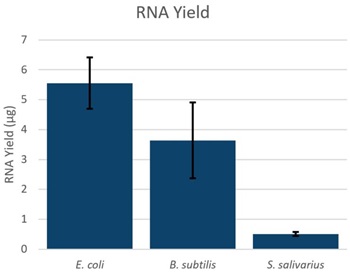
Escherichia coli, Bacillus subtilis and Streptococcus saliverius were grown overnight in LB media at 37 °C. The E. coli culture grew to an OD of 6.59, the B. subtilis culture grew to an OD of 4.15, and the S. saliverius culture grew to an OD of 0.94. RNA was extracted from 150 µL of those overnight cultures. Yield was measured by absorbance at 260nm on a NanoDrop (Thermo Fisher Scientific), and integrity was assessed on an Agilent RNA ScreenTape (Agilent). The yield of RNA correlated to the bacterial overnight OD, with higher overnight OD resulting in higher yields (Figure 1).
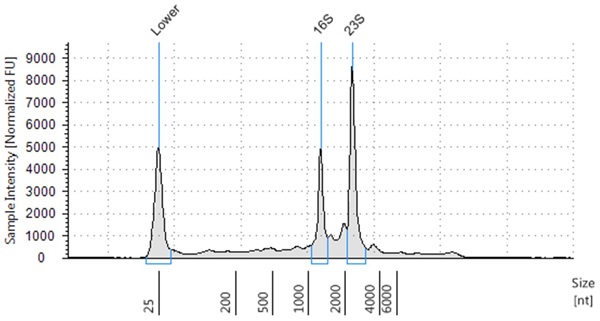
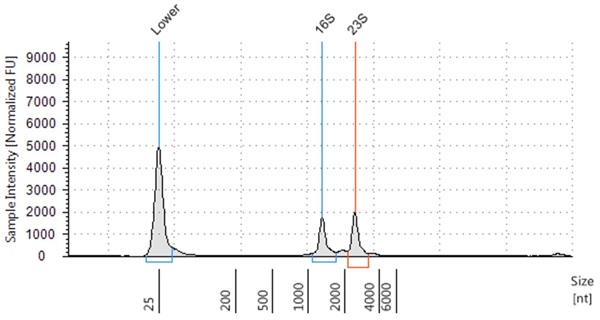
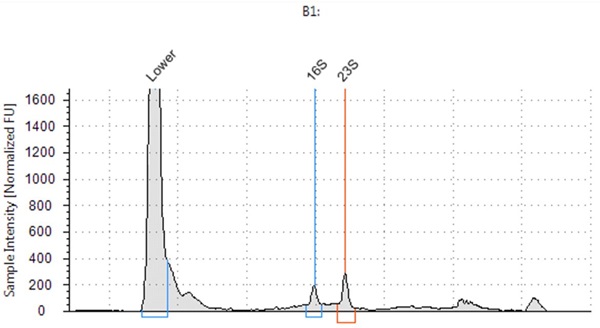
The resulting RNA extracted had high RIN scores, with a RIN score of 8.4 for E. coli, 9.4 for B. subtilis and 7.4 for S. saliverius (Figure 2-4) indicating highly intact RNA.